Not just authors of Greek and Roman medical literature but also a number historians have provided surprisingly elaborate descriptions of (infectious) disease in antiquity. Potentially one of the most prominent examples is the description of the Plague of Athens in Thucydides History of the Peloponnesian War [1]. Thucydides describes the plague as having originated in northeastern Africa, spreading from Egypt and Lybia to Minor Asia and Greece, ultimately reaching Athens from Piraeus (2:48) which interestingly is also a likely path for the emergence of falciparum malaria in the Mediterranean (see below). For his time (and the fact that he had no medical training) Thucydides demonstrates impressive insight into basic epidemiology (e.g. that recovery from the disease provided a high level of protection from new infection and that the disease also occurs in animals). However, although his description of the symptoms (2:49) is rather detailed (largely also due to the fact that he had suffered from the disease himself) it is virtually impossible to associate these with any current infectious disease (many of which, however, tend to have rather uncharacteristic clinical presentations).

This also leads to the question of causality. Malaria is commonly associated with poverty (i.e. is a so called Disease of Poverty). It forms the core of a vicious circle of disease and poverty: the disease causes poverty, which then leads to a further spread of disease. Applied to the context of Mediterranean history: was the spread of a new and more deadly form of malaria instrumental in the decline of ancient civilizations in late antiquity or was the spread of malaria a consequence of declining civilizations? In how far falciparum malaria contributed to or emerged secondary to the decline and finally fall of the Roman Empire remains a matter of discussion [8]. Both theories, namely the fact that it was one of the causes of the decline, or that it developed and spread following the deterioration of infrastructure in late antiquity, seem rather plausible. However, there is virtually no physical evidence to prove either.
Antigen detection
 Over the past centuries the Plague of Athens (fig. 1) has therefore been associated with pretty much any imaginable bacterial, viral, and rickettsial infectious disease ranging from typhus to typhoid, to influenza and hemorrhagic fevers. A large scale investigation published in 2006 using dental samples originating from a mass burial site (fig. 2) just outside of Kerameikos cemetry excavated in the 1990s by E. Baziotopoulou-Valavani [2] tried to shed new light on a discussion that had been going on for centuries. M.J. Papagrigorakis et al. [3] concluded based on state of the art diagnostic methodology that the likely cause of the Plague of Athens had been typhoid fever. However, this view has been challenged by a number of authors, including B. Shapiro et al. [4] who conclude based on phylogenetic analysis that contamination is a likely cause for the misidentification of typhoid as the cause of the Plague. From a modern perspective and based on our extensive experience with a variety of hemorrhagic fevers, many of which have their animal reservoirs in Africa, the described symptoms would likely best match this group of diseases. However, identification of viral genetic footprints remains extremely difficult, demonstrating challenges and inherent limitations of even the most advanced diagnostic technologies in paleopathology.
Over the past centuries the Plague of Athens (fig. 1) has therefore been associated with pretty much any imaginable bacterial, viral, and rickettsial infectious disease ranging from typhus to typhoid, to influenza and hemorrhagic fevers. A large scale investigation published in 2006 using dental samples originating from a mass burial site (fig. 2) just outside of Kerameikos cemetry excavated in the 1990s by E. Baziotopoulou-Valavani [2] tried to shed new light on a discussion that had been going on for centuries. M.J. Papagrigorakis et al. [3] concluded based on state of the art diagnostic methodology that the likely cause of the Plague of Athens had been typhoid fever. However, this view has been challenged by a number of authors, including B. Shapiro et al. [4] who conclude based on phylogenetic analysis that contamination is a likely cause for the misidentification of typhoid as the cause of the Plague. From a modern perspective and based on our extensive experience with a variety of hemorrhagic fevers, many of which have their animal reservoirs in Africa, the described symptoms would likely best match this group of diseases. However, identification of viral genetic footprints remains extremely difficult, demonstrating challenges and inherent limitations of even the most advanced diagnostic technologies in paleopathology.
Paleopathology (i.e. the study of disease and injuries in human remains such as skeletal or mummified tissue) has provided evidence of human disease and is essential for understanding the impact of disease on human history [5]. It uses an interdisciplinary approach to identify causes of disease and injuries based on e.g. archaeological, anthropological, medical, or epidemiological evidence. Traditionally paleopathology had to rely on identifying morphological changes and lesions (mostly on bones and teeth), which are typically associated with chronic disease, either directly visible or identified with the help of X-ray, CT, microscopy etc. However, many diseases and particularly acute infectious diseases leave virtually no traces on skeletal remains. Here the past 25 years have brought major advances that are largely based on progress made in the medical diagnostic field and modification of these techniques to the specific needs of paleopathology. This has resulted in new insights into the history of acute diseases such as malaria, which often kill their host before leaving visible traces on bones or teeth.
Although malaria remains a major public health threat throughout large parts of the world, its origins and spread as well as its impact on ancient civilizations remain poorly understood and the role malaria played in the history of Mediterranean civilizations (fig. 3) has been discussed controversially. In large parts this is due to the fact that we still do not fully understand when and how Plasmodium falciparum, undoubtedly the most deadly of the four human malaria parasites reached the Mediterranean. Phylogenetic evidence suggests that P. falciparum populations have evolved from Laverania species, a parasite found in gorillas in Africa [6]. However, there is no consensus on when falciparum malaria finally reached Mediterranean and European civilizations [7]. It is generally acknowledged that P. falciparum is the youngest of the four human malaria parasites and that it is likely that it reached the northern Mediterranean rather late in antiquity. On the one hand it tends to kill its host making it a rather poorly adapted (and therefore young) parasite in humans. On the other hand it has been argued that in the presence of a disease as deadly and devastating as falciparum malaria the Mediterranean cultures of antiquity could never have thrived.

Morphological identification of skeletal changes
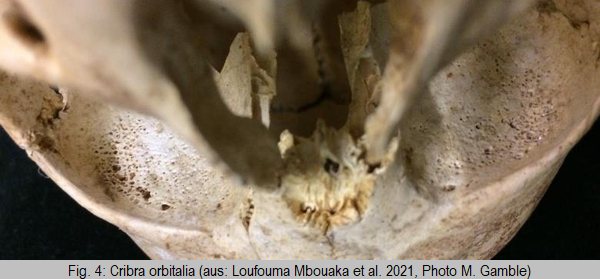
Porotic hyperosteosis and cribra orbitalia (fig. 4) have been used as proxies to identify malaria in skeletal remains for decades [9]. This is based on the assumption that malaria is commonly associated with chronic anemia. To a certain extent this is correct and works through two different mechanisms. First, chronic malaria is associated with chronic anemia which is reflected in the aforementioned skeletal changes. Second, malaria seems to play a role in selecting for erythrocytic (i.e. red blood cell) disorders, which may confer a certain level of protection from severe disease while at the same time leading to chronic anemia. However, although the presence of such erythrocytic disorders may be an indicator of the presence of malaria within a certain population, it has no relation to malaria infection in a given individual. Similarly, most individuals dying of malaria (today predominantly children [10]) die of acute disease rather than chronic infections, which will not lead to any identifiable skeletal lesions. Skeletal changes may therefore be helpful in identifying the presence of malaria in ancient populations but may be of very limited use for identifying malaria as a cause of death.
In modern medicine immunoassays are widely used to identify disease-specific antigens (or antibodies). Antigens commonly used as markers of malaria infection are histidine-rich-protein II (HRP2), an abundant and highly stable protein specific to P. falciparum [11], plasmodium lactate dehydrogenase (pLDH), and aldolase. By far the most commonly used immunoassays to identify malaria are the so called immunochromatographic assays, which allow for simple, economical field-deployable assays that require little training and can be conducted without any lab infrastructure (lateral flow assays or RDTs). Here the antigen forms a complex with a conjugated (colored) antibody, is transported along a nitrocellulose membrane and finally captured by a second antibody coated to a test line resulting in a visible color change. These assays are widely commercially available and very easy to use and have e.g. been used to identify malaria in Egyptian mummies [12] as well as bone samples from the Medici Family [13]. However, they are specifically designed to be used on biological (blood) samples (which do not include any potential contaminants, e.g. originating from soil) [14]. Although the sensitivity in modern RDTs is high, their specificity (i.e. the question of potential false positives) in ancient samples needs further validation. Enzyme-linked immunosorbent assays (ELISAs) use a very similar approach (using two antibodies, the first coated to a plate, the second conjugated to an enzyme which can be detected as a color change using a substrate) but require considerably more lab infrastructure and training [15]. However, although more difficult to establish the format allows for more flexibility and ultimately for establishing ELISA protocols that are specifically adapted to the needs of paleopathology thereby overcoming the specificity issues associated with RDTs.
Molecular identification of disease in ancient samples
Ancient DNA (aDNA) has typically undergone a process of degradation and is therefore considerably more difficult to analyse than contemporary biological samples. Under optimal condition DNA can survive for thousands of years, but temperature, humidity, pH and other environmental factors have a major impact on the degradation of DNA through fragmentation/depurination and deamination [16]. Pulp and dentine found in teeth are best suited to preserve aDNA and are therefore most commonly used as a sample source. Polymerase chain reaction (PCR) has traditionally been the most commonly employed method for analyzing aDNA samples. Originally primers targeting the plasmodial 18S rRNA, a technique commonly used in malaria diagnosis, were used for molecular identification of malaria in ancient samples. This has now largely been replaced by capture-enrichment and sequencing of the mitochondrial DNA of the parasite. Next generation sequencing (NGS) has revolutionized aDNA research. A shotgun approach (a technique in which DNA is broken up randomly into numerous small segments, which are then sequenced and reassembled using computer algorythms based on multiple overlaps between fragments) is well suited for aDNA, which is typically already highly fragmented. Another NGS technique employed is using a capture assay targeting the mitochondrial Plasmodium genome (mtDNA - mitochondria are organelles carrying multiple copies of their own genetic material), which is particularly suited for testing also very small DNA fragments [17].
Conclusions
The identification of pathogens in ancient human remains has seen major advances in the past 25 years. However, the analysis of highly degraded material, as commonly found in ancient samples, remains extremely challenging as both highly fragmented genetic material as well as degraded proteins are difficult to identify. In spite of this progress all currently used techniques have their inherent limitations. A combined approach comprising different methodologies is therefore highly indicated [18].
Acknowledgements
I wish to thank A. Loufouma Mbouaka, PhD student at the Medical University of Vienna, M. Gamble and M. Binder from the Austrian Archaeological Institute, and our collaborators from the Institute for Mummy Studies in Bolzano, Italy for our collaboration on this topic.
References
Angel 1966
J.L. Angel, Porotic hyperostosis, anemias, malarias, and marshes in the prehistoric Eastern Mediterranean, Science 153(3737), 1966, 760763
Bianucci 2008
R. Bianucci G. Mattutino R. Lallo P. Charlier H. Jouin-Spriet A. Peluso T. Higham C. Torre ER. Massa. Immunological evidence of Plasmodiumfalciparum infection in an Egyptian child mummy from the Early Dynastic Period. Journal of Archaeological Science, 35(7), 2008, 1880-1885
Bruce-Chwatt de Zulueta 1980
L. Bruce-Chwatt J. de Zulueta, The Rise and Fall of Malaria in Europe: A Historico-Epidemiological Study (Oxford 1980)
Dabney et al. 2013
J. Dabney M. Meyer S. Pδδbo, Ancient DNA Damage, Cold Spring Harbor Perspectives in Biology 2013, Jul. 5(7), doi: 10.1101/cshperspect.a012567
Fornaciari 2010
G. Fornaciari V. Giuffra E. Ferroglio S. Gino R. Bianucci. Plasmodium falciparum immunodetection in bone remains of members of the Renaissance Medici family (Florence, Italy, sixteenth century). Transactions of the Royal Society of Tropical Medicine and Hygiene, 104(9), 2010, 583587
Kiesslich 1998
J. Kiesslich, Ancient DNA, Forum Archaeologiae 9/12/98 (accessed 23 July 2021)
Liu et al. 2010
W. Liu V. Li G.H. Learn R.S. Rudicell J.D. Robertson B.F. Keele J.B. Ndjango C.M. Sanz et al., Origin of the human malaria parasite Plasmodium falciparum in gorillas, Nature 467 (7314), 2010, 420425
Loufouma Mbouaka et al. 2020
A. Loufouma Mbouaka M. Binder H. Noedl M. Gamble, Evaluation of rapid diagnostic tests and Enzyme Linked Immunoassay in the detection of malaria in ancient human remains, Journal of Archaeological Science 116, 2020, 105118
Loufouma Mbouaka et al. 2021
A. Loufouma Mbouaka M. Gamble C. Wurst H. Yoko Jδger F. Maixner A. Zink H. Noedl M. Binder, The elusive parasite: comparing macroscopic, immunological, and genomic approaches to identifying malaria in human skeletal remains from Sayala, Egypt (third to sixth centuries AD), Archaeological and Anthropological Sciences 13 (7), 2021, 115
Merheb et al. 2019
M. Merheb R. Matar R. Hodeify S.S. Siddiqui C.J. Vazhappilly J. Marton S. Azharuddin H.A.L. Zouabi, Mitochondrial DNA, a Powerful Tool to Decipher Ancient Human Civilization from Domestication to Music, and to Uncover Historical Murder Cases, Cells 8 (5), 2019, 433
Noedl et al. 2002
H. Noedl C. Wongsrichanalai R.S. Miller K.S. Myint S. Looareesuwan Y. Sukthana V. Wongchotigul H. Kollaritsch G. Wiedermann W.H. Wernsdorfer, Plasmodium falciparum: effect of anti-malarial drugs on the production and secretion characteristics of histidine-rich protein II, Experimental Parasitology. 2002 Nov-Dec 102 (3-4), 2002, 157163
Noedl et al. 2006
H. Noedl K. Yingyuen A. Laoboonchai M. Fukuda J. Sirichaisinthop R.S. Miller, Sensitivity and specificity of an antigen detection ELISA for malaria diagnosis, American Journal of Tropical Medicine and Hygiene Dec 75 (6), 2006, 12051208
Papagrigorakis et al. 2006
M.J. Papagrigorakis C. Yapijakis P.N. Synodinos E. Baziotopoulou-Valavani, DNA examination of ancient dental pulp incriminates typhoid fever as a probable cause of the Plague of Athens, International Journal of Infectious Diseases 10 (3), 2006, 206214
Parlama Stampolidis 2000
L. Parlama N.Ch. Stampolidis, The City beneath the City (Athen 2000)
Sallares Bouwman Anderung 2004
R. Sallares A. Bouwman C. Anderung, The spread of malaria to Southern Europe in antiquity: new approaches to old problems, Medical History Jul 48 (3), 2004, 311328, doi: 10.1017/s0025727300007651
Shapiro et al. 2006
B. Shapiro A. Rambaut M. Gilbert P. Thomas et al., No proof that typhoid caused the Plague of Athens (a reply to Papagrigorakis et al.), International Journal of Infectious Diseases 10 (4), 2006, 334335
Thucydides: History of the Peloponnesian War. https://en.wikisource.org/wiki/History_of_the_Peloponnesian_War/Book_2 (accessed 23 June 2021)
World Malaria Report 2020
WHO Global Malaria Programme (Geneva 2020, ISBN 978-92-4-001579-1)
[1] Thucydides: History of the Peloponnesian War, book 2.
[2] E. Baziotopoulou-Valavani I. Tsirigoti-Drakotou in: Parlama Stampolidis 2000, 272.
[3] Papagrigorakis et al. 2006.
[4] Shapiro et al. 2006.
[5] Kiesslich 1998.
[6] Liu et al. 2010.
[7] Sallares Bouwman Anderung 2004.
[8] Bruce-Chwatt de Zulueta 1980.
[9] Angel 1966.
[10] World Malaria Report 2020.
[11] Noedl et al. 2002.
[12] Bianucci et al. 2008.
[13] Fornaciari et al. 2010.
[14] Loufouma Mbouaka et al. 2020.
[15] Noedl et al. 2006.
[16] Dabney et al. 2013.
[17] Merheb et al. 2019.
[18] Loufouma Mbouaka et al. 2021.
![]() © Harald Noedl
© Harald Noedl
e-mail: noedlh@gmail.com
This article should be cited like this: H. Noedl, Tracing infectious disease pathogens in ancient human remains. 25 years of advances in paleopathology, Forum Archaeologiae 100/IX/2021 (http://farch.net).